When I was a grad student, I felt the immense pain to "type" every single citation for my dissertation and many drafts of my manuscripts. Given that there are 4.5 million doctorate degree holders, I was surprised that no citation generater feature is available in the widely-used PubMed.
I therefore wrote a Python program that accesses NCBI’s PubMed database (via Entrez API) by which after finding and retrieving article information, MLA citations are generated.
This script runs quickly; this is the beauty of automation!
Demo
In this tutorial, I will provide step-by-step instructions in writing the Python script to do so! Good luck!
Prerequisites
- Familiarity of Python (data structures, functions, indexing)
- Basic knowledge of web API
- Visual Studio Code - but any code editer works
Getting started
Install requests, a Python module as described here. This will allow your script to retrieve information.
Open your terminal and let's create a directory (folder) called citation-generator. Go into the directory.
$ mkdir citation-generator $ cd citation-generater
Create a new file called main.py and open the file with the code editor (the open command will open the file using your default editor).
$ touch main.py $ open main.py
In the code editor with main.py opened, it is convention to import all modules or dependences that the script needs to run.
Let's start coding!
First, create a function called pubmed_search with an argument called s. The argument is an input that must be passed into the function so that the function can use the argument to "do something" or use the argument as a parameter when calling the function.
NCBI's databases are openly available using their "base url", as described in their documentation. We will include this link in the base_url as a string in the pubmed_search function, along with the database of choice (pubmed) as a string.
# in main.py
import requests
import json
def pubmed_search(s):
base_url = "https://eutils.ncbi.nlm.nih.gov/entrez/eutils/"
database = "pubmed"
So how does the script find information in NCBI? From the NCBI's Entrez documentation, the following link can help navigate to the NCBI API page using an example search term: big data etl.
https://eutils.ncbi.nlm.nih.gov/entrez/eutils/esearch.fcgi?db=pubmed&term=big+data+etl
The page link returns a response in XML format! It provides the search result count and list of PubMed article IDs - which we will need to generate our citations.
Let's create a variable called query_url to build that search link.
# in main.py
import requests
import json
def pubmed_search(s):
base_url = "https://eutils.ncbi.nlm.nih.gov/entrez/eutils/"
database = "pubmed"
query_url = base_url + "esearch.fcgi?db=" + database + "&term=" + s + "&retmode=json"
Notice the "retmode" at the end of query_url. We want the query to return the information in JSON instead of in XML format, which is almost like a Python dictionary, as shown in https://eutils.ncbi.nlm.nih.gov/entrez/eutils/esearch.fcgi?db=pubmed&term=big+data+etl&retmode=json.
For Python to retrieve this information, we use the requests.get() method and call the string “response”. We need to build another search query for each article listed in the ID list (which is conveniently stored as a list) and just like in Python dictionaries, we can access this ID list by referring to the “key” into a new string called “idList”. This is why the JSON format is helpful – Python can parse data from JSON as if JSON is a dictionary!
# in main.py
import requests
import json
def pubmed_search(s):
base_url = "https://eutils.ncbi.nlm.nih.gov/entrez/eutils/"
database = "pubmed"
query_url = base_url + "esearch.fcgi?db=" + database + "&term=" + s + "&retmode=json"
response = requests.get(query_url)
idList = response["esearchresult"]["idlist"]
Let’s create an if else conditional when there are no numbers under idList using the len() function, we print a message. As for else, we create a for loop to iterate the first 5 articles (PubMed allows for a maximum of 20 articles).
We would like to keep track of how many articles are listed (from 1 to 5) so let’s create a new string called “total” and check the count for each article as Python performs the search by creating a new string called “i” and using the .index() function.
Python indexing starts at 0 so we need to create a new variable called article_count that adds 1 to the index.
# in main.py
import requests
import json
def pubmed_search(s):
base_url = "https://eutils.ncbi.nlm.nih.gov/entrez/eutils/"
database = "pubmed"
query_url = base_url + "esearch.fcgi?db=" + database + "&term=" + s + "&retmode=json"
response = requests.get(query_url)
idList = response["esearchresult"]["idlist"]
if len(idList) == "0":
print("No articles found! Please try another search term.")
else:
for article in idList[0:5]:
total = len(idList[0:5])
i = idList.index(article)
article_count = int(i) + 1
Let’s have Python print a message when it begins a search so create a conditional when the index starts at 0. To track the search of each article, we are going to print a message for the count number of each article out of the total.
# in main.py
import requests
import json
def pubmed_search(s):
base_url = "https://eutils.ncbi.nlm.nih.gov/entrez/eutils/"
database = "pubmed"
query_url = base_url + "esearch.fcgi?db=" + database + "&term=" + s + "&retmode=json"
response = requests.get(query_url)
idList = response["esearchresult"]["idlist"]
if len(idList) == "0":
print("No articles found! Please try another search term.")
else:
for article in idList[0:5]:
total = len(idList[0:5])
i = idList.index(article)
article_count = int(i) + 1
if i == "0":
print("--------------")
print("Beginning article search")
print("--------------")
print(f"Retrieving articles {article_count} of {total}")
If we want to access the information for the first article of our example search, we use NCBI’s “esummary” and the ID 31201587: https://eutils.ncbi.nlm.nih.gov/entrez/eutils/esummary.fcgi?db=pubmed&id=31201587&retmode=json. Once clicked, you can see a wealth of information of the article, including the article title, source, each author, and publication date. This is what we need to build our citation.
Just as we did earlier, we need to create another base url link called url, and the variable search_url for each “article” listed in idList in this for loop. We also need to create a new variable called search_response (in JSON format) to store our requests from the response by search_url.
# in main.py
import requests
import json
def pubmed_search(s):
base_url = "https://eutils.ncbi.nlm.nih.gov/entrez/eutils/"
database = "pubmed"
query_url = base_url + "esearch.fcgi?db=" + database + "&term=" + s + "&retmode=json"
response = requests.get(query_url)
idList = response["esearchresult"]["idlist"]
if len(idList) == "0":
print("No articles found! Please try another search term.")
else:
for article in idList[0:5]:
total = len(idList[0:5])
i = idList.index(article)
article_count = int(i) + 1
if i == "0":
print("--------------")
print("Beginning article search")
print("--------------")
print(f"Retrieving articles {article_count} of {total}")
url = "https://eutils.ncbi.nlm.nih.gov/entrez/eutils/esummary.fcgi?"
search_url = url + "db=" + database + "&id=" + article + "&retmode=json"
search_response = requests.get(search_url).json()
In our example article response for ID 31201587, notice how there are multiple authors listed under “authors” and “name”. We need to create an empty list called article_list to store each “name” (we will iterate each name in a new for loop later).
To keep the script running in case there is no information listed for the article, we will add a try execpt loop.
For creating citations of each article, we want to know the authors, title, journal, year, volume, issue, pages, and doi. Just like a Python dictionary, we can parse the data by accessing the values of each of these “keys” that we want and creating a new string for each.
Let’s have Python print a message when it begins a search so create a conditional when the index starts at 0. To track the search of each article, we are going to print a message for the count number of each article out of the total.
# in main.py
import requests
import json
def pubmed_search(s):
base_url = "https://eutils.ncbi.nlm.nih.gov/entrez/eutils/"
database = "pubmed"
query_url = base_url + "esearch.fcgi?db=" + database + "&term=" + s + "&retmode=json"
response = requests.get(query_url)
idList = response["esearchresult"]["idlist"]
if len(idList) == "0":
print("No articles found! Please try another search term.")
else:
for article in idList[0:5]:
total = len(idList[0:5])
i = idList.index(article)
article_count = int(i) + 1
if i == "0":
print("--------------")
print("Beginning article search")
print("--------------")
print(f"Retrieving articles {article_count} of {total}")
url = "https://eutils.ncbi.nlm.nih.gov/entrez/eutils/esummary.fcgi?"
search_url = url + "db=" + database + "&id=" + article + "&retmode=json"
search_response = requests.get(search_url).json()
article_list = []
try:
pubmed_id = search_response["result"][article]["uid"]
title = search_response["result"][article]["title"]
authors = search_response["result"][article]["authors"]
journal = search_response["result"][article]["source"]
pub_date = search_response["result"][article]["pubdate"]
volume = search_response["result"][article]["volume"]
issue = search_response["result"][article]["issue"]
pages = search_response["result"][article]["pages"]
doi = search_response["result"][article]["elocationid"]
except:
print("No information found. Skipping...")
Because the string “author” contains multiple values, we append these values to our empty “authors_list” using a new for loop, and we use the replace() method to remove ‘, [, ] characters when printing the results in terminal.
Additionally, for the article title, we also need to replace <i> (which denotes as italics in XML) to and (without the spacing), which lets the user know that the word is italicized.
Finally, we can print out our results in terminal by showing the PubMed ID and place the strings in citation format. Note that we use indexing to just print the year. We are almost done!
# in main.py
import requests
import json
def pubmed_search(s):
base_url = "https://eutils.ncbi.nlm.nih.gov/entrez/eutils/"
database = "pubmed"
query_url = base_url + "esearch.fcgi?db=" + database + "&term=" + s + "&retmode=json"
response = requests.get(query_url)
idList = response["esearchresult"]["idlist"]
if len(idList) == "0":
print("No articles found! Please try another search term.")
else:
for article in idList[0:5]:
total = len(idList[0:5])
i = idList.index(article)
article_count = int(i) + 1
if i == "0":
print("--------------")
print("Beginning article search")
print("--------------")
print(f"Retrieving articles {article_count} of {total}")
url = "https://eutils.ncbi.nlm.nih.gov/entrez/eutils/esummary.fcgi?"
search_url = url + "db=" + database + "&id=" + article + "&retmode=json"
search_response = requests.get(search_url).json()
article_list = []
try:
pubmed_id = search_response["result"][article]["uid"]
title = search_response["result"][article]["title"]
authors = search_response["result"][article]["authors"]
journal = search_response["result"][article]["source"]
pub_date = search_response["result"][article]["pubdate"]
volume = search_response["result"][article]["volume"]
issue = search_response["result"][article]["issue"]
pages = search_response["result"][article]["pages"]
doi = search_response["result"][article]["elocationid"]
for i in authors:
all_authors = i["name"]
author_list.append(all_authors)
names = str(author_list).replace("'", "").replace("[","").replace("]","")
# please see this line on Github, code is not displaying properly
corrected_title = title.replace("<i>", "").replace("</i>", "")
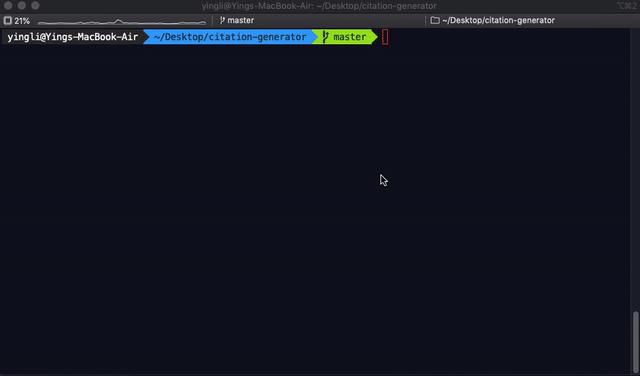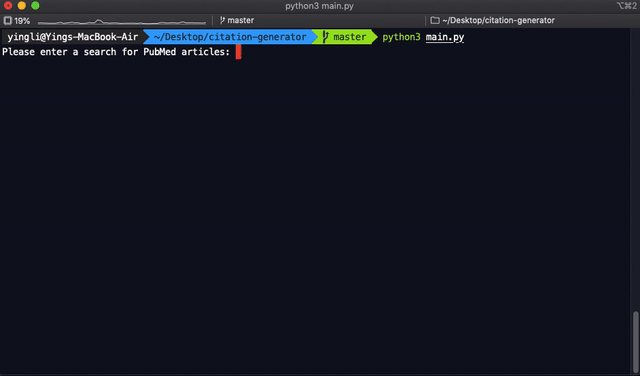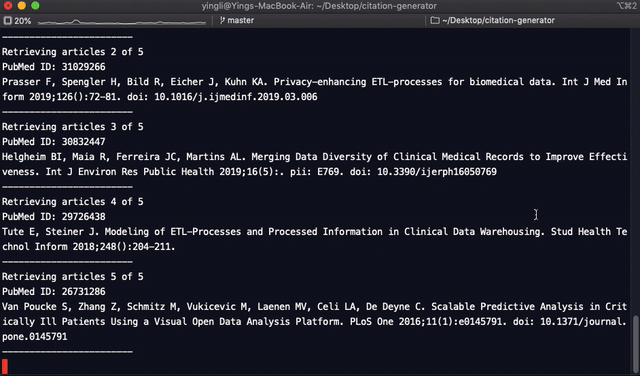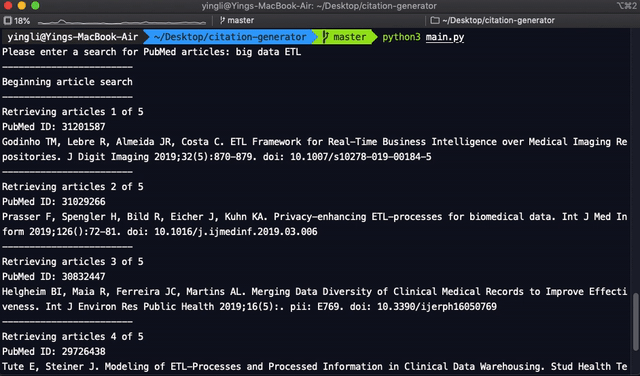
print(f"PubMed ID: {pubmed_id}")
print(f"{names}.{corrected_title} {journal} {pub_date[0:4]};{volume}({issue}):{pages}. {doi}")
print("-----------------")
except:
print("No information found. Skipping...")
We are done with writing our function so outside of the function, we create a string called “term” to take in user’s input for the search terms and call the function. Each spacing in the term must be replaced by a “+” sign so we use the replace() function again.
# in main.py
import requests
import json
def pubmed_search(s):
### CODE BLOCK ###
term = input("Please enter a search for PubMed articles :").replace(" ", "+")
pubmed_search(term)
Congrats, the script is done! You can refer to the code here.